What do the colours on the pathway heatmaps represent? - FastBMD - OmicsForum
The pathway heatmap is an appealing visualization to clearly shows how the expression of each pathway gene compares to the others. It is generated when you click a pathway or gene set name in the “Gene Set Enrichment” panel at the result page. An example output is shown below The pathway heatmap values are calculated through a series of steps: The fitted model for each gene is evaluated across the range of doses in the uploaded data. The resulting modeled expression values are normalized

Heatmap based on the main KEGG pathways predicted at the different
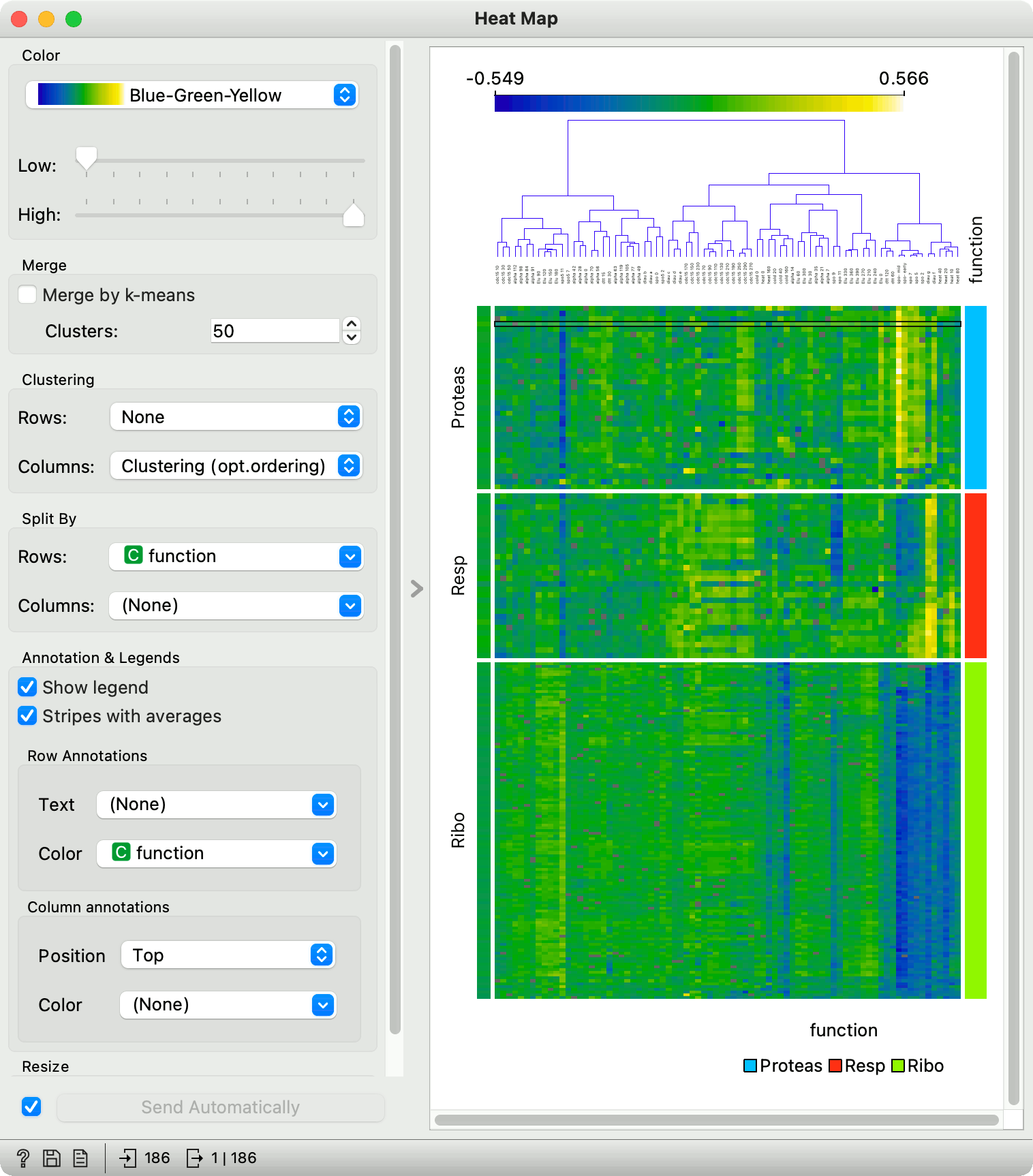
Heat Map — Orange Visual Programming 3 documentation
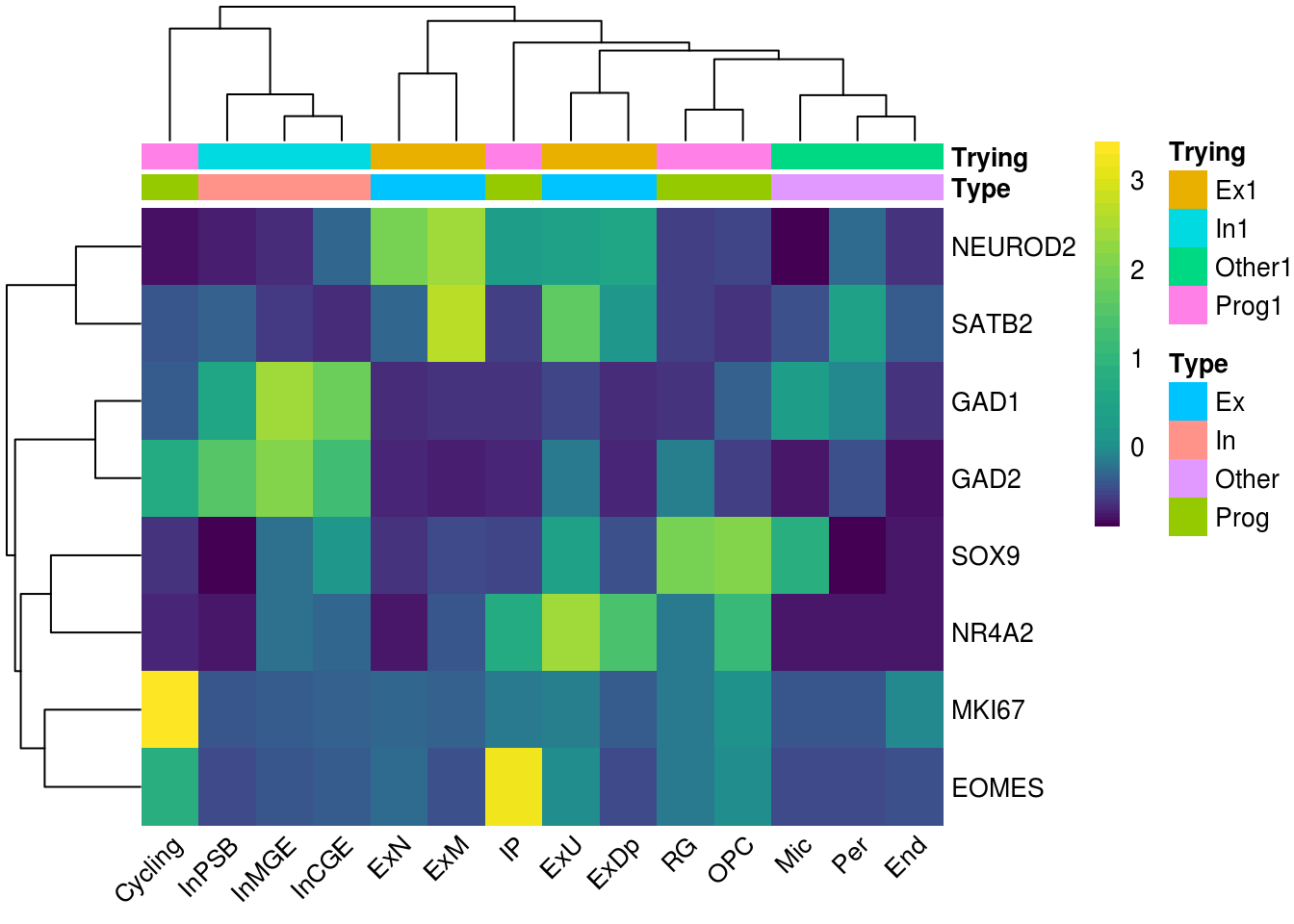
Chapter 9 Heatmap Color Palette Single Cell Multi-Omics Data Analysis
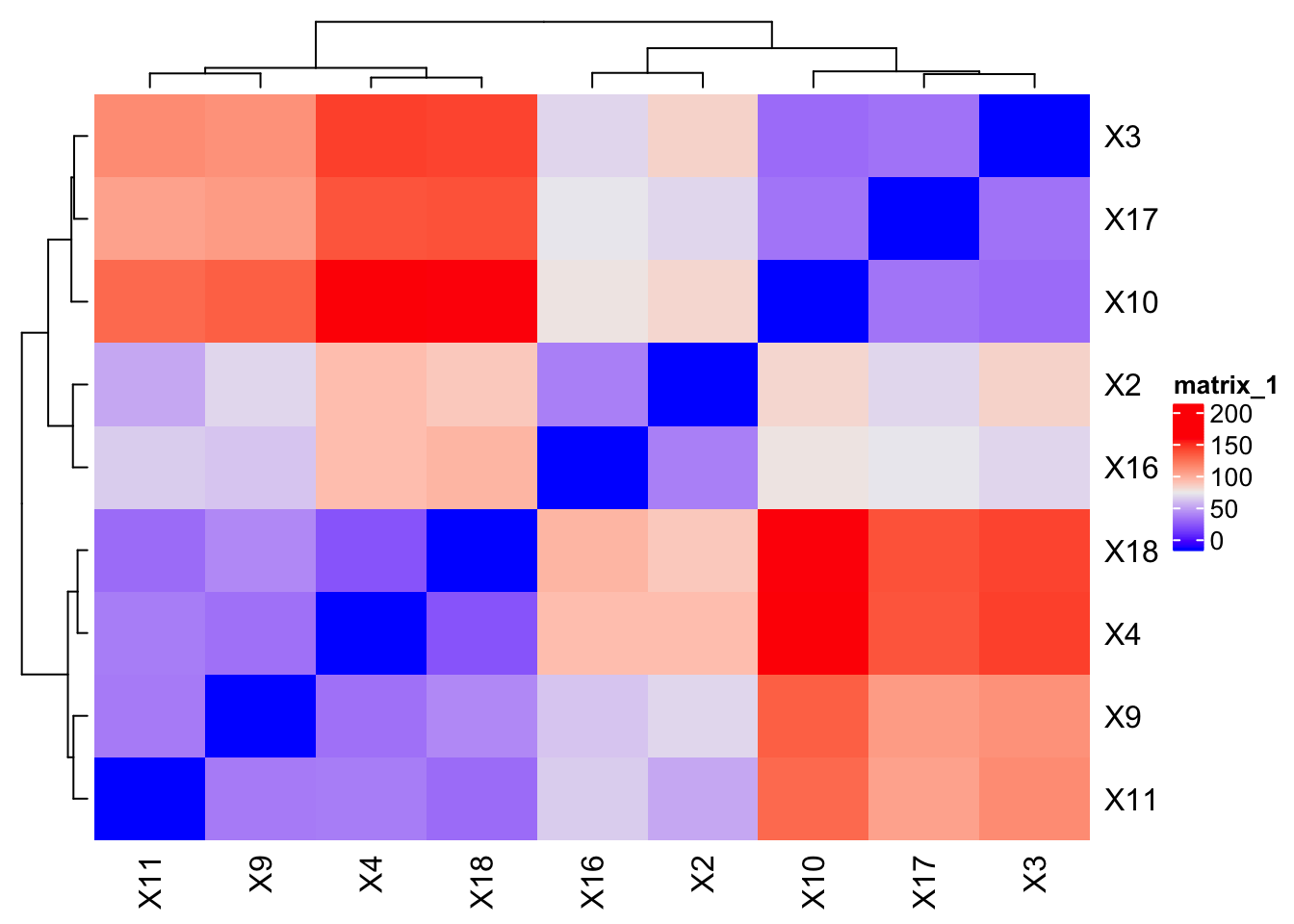
Let's Plot 6: Simple guide to heatmaps with ComplexHeatmaps
KEGG Decoder heat map based on Kofamscan results. The heat map
Bitesize Bioinformatics: Drawing Heatmaps in R

A Heatmap of-log10-transformed P-values for all 32 significant CP:KEGG
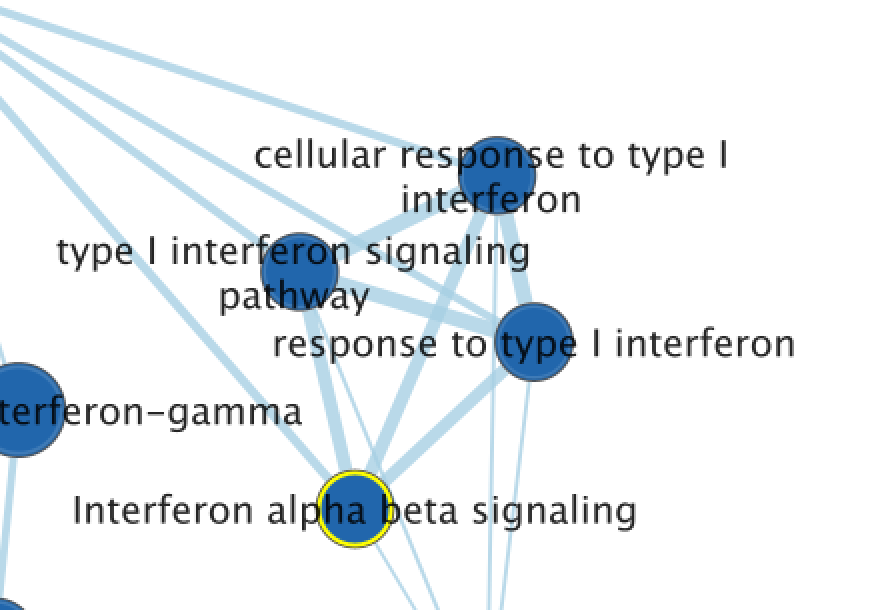
13A) Exploring the Table Panel heat map Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap
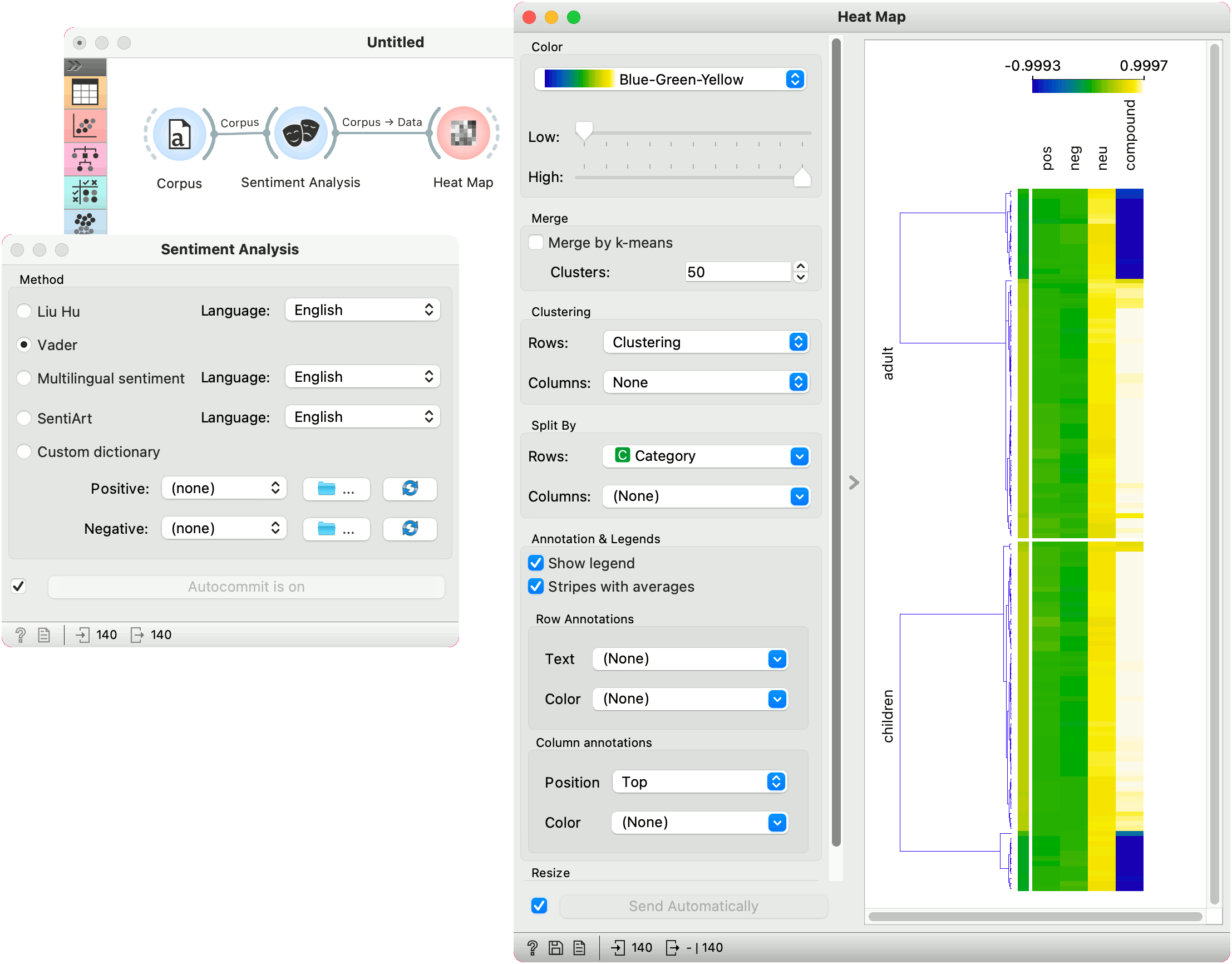
Heat Map — Orange Visual Programming 3 documentation

Smooth velocity fields for tracking climate change

Heatmap showing the conservation of individual metabolic pathways. Each
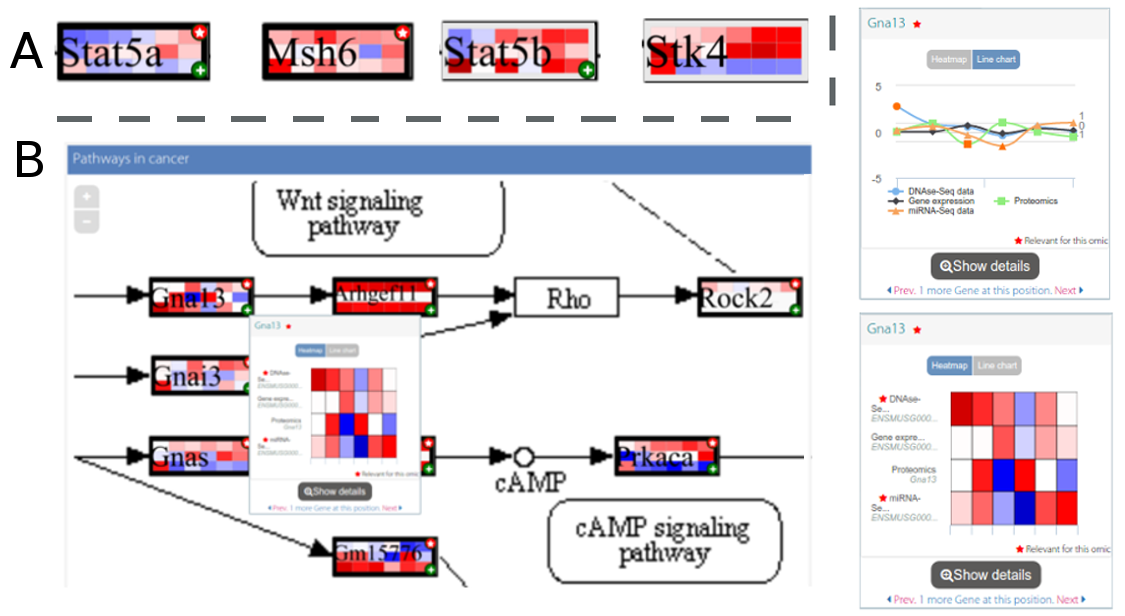
Multi-omic pathway-based visualisation - Paintomics Documentation

How to interpret heatmap ?
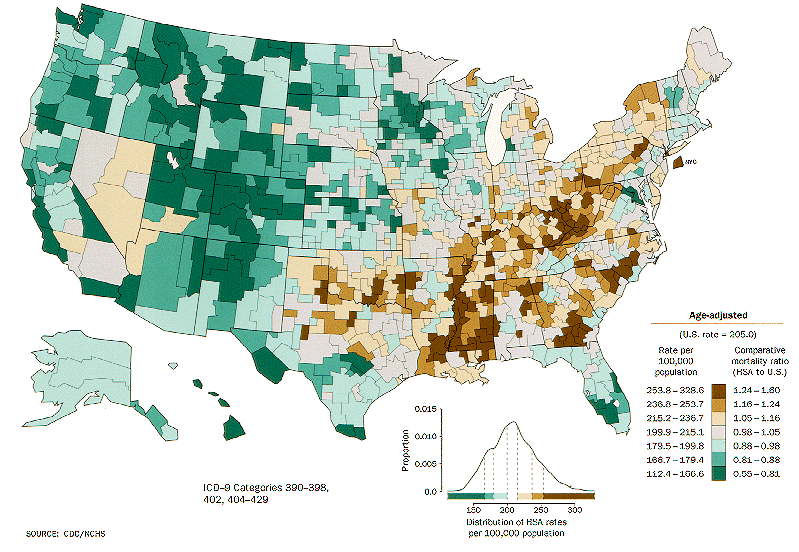
How to choose colors for maps and heat maps - The DO Loop
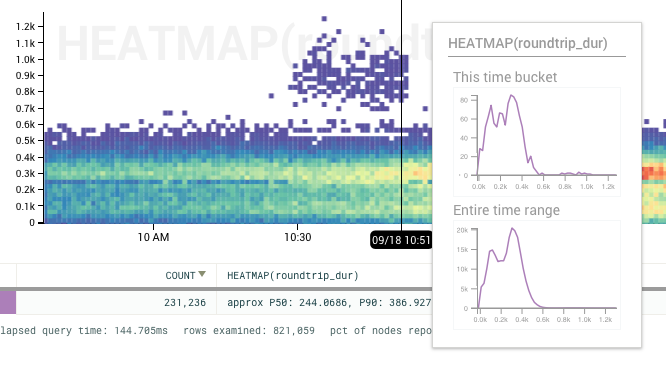
Heatmaps Are The New Hotness*