Genotyping by low-coverage whole-genome sequencing in intercross
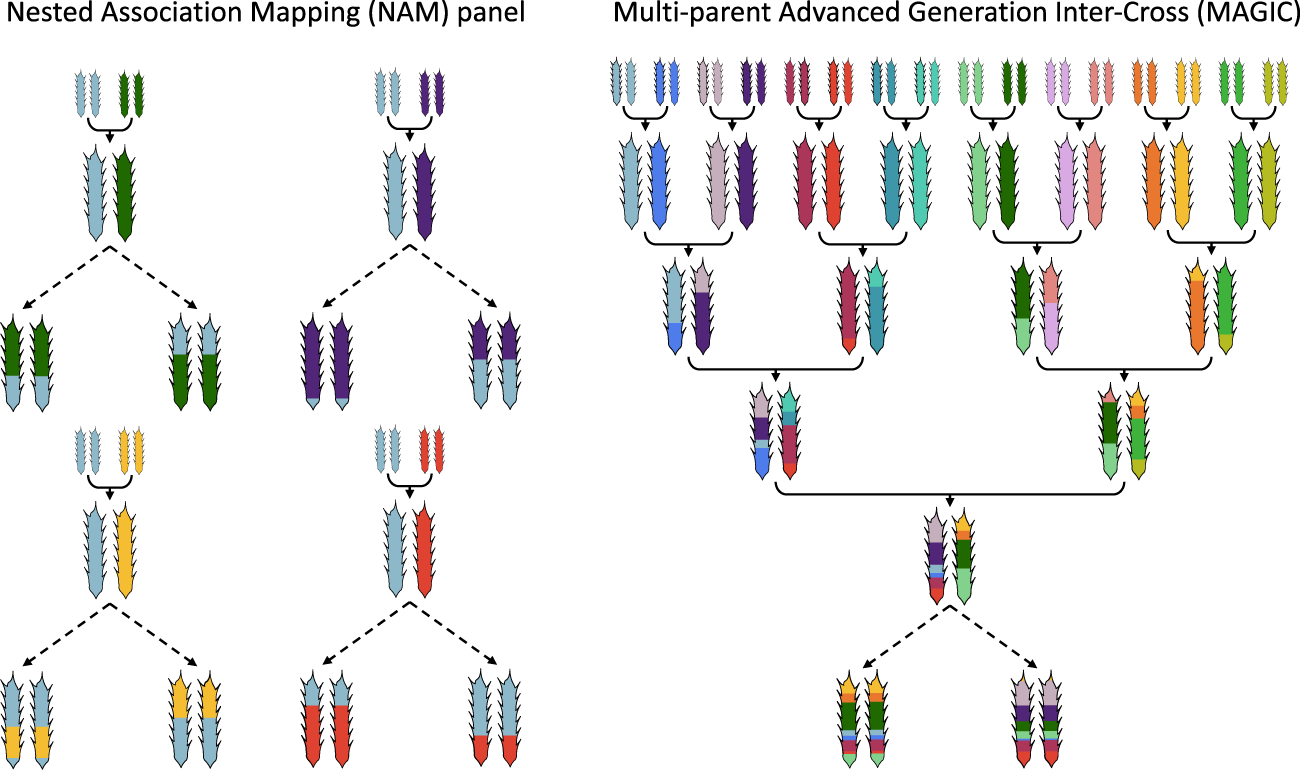
Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding
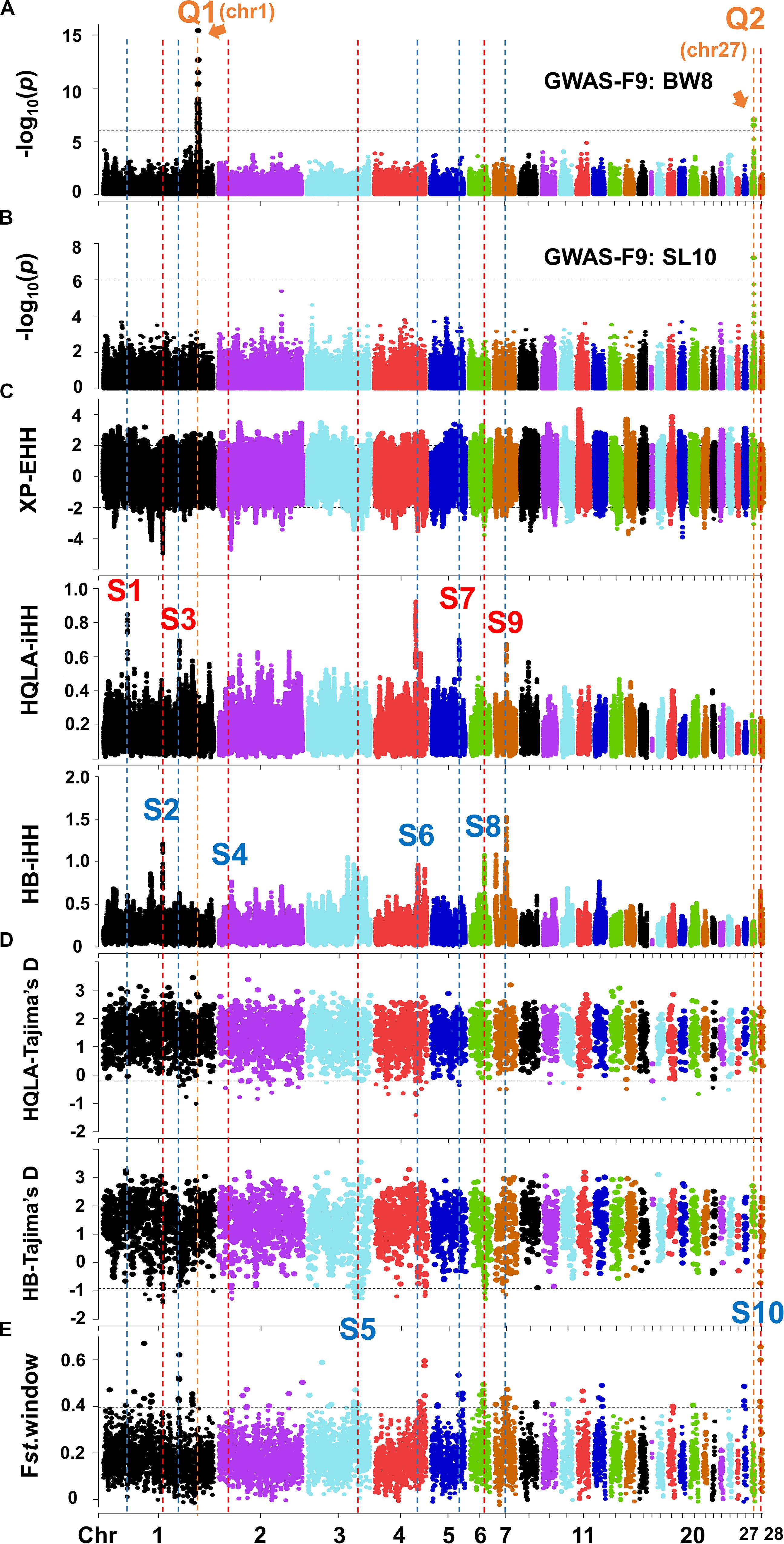
Frontiers Genetic Dissection of Growth Traits in a Unique Chicken Advanced Intercross Line
Diversity, Free Full-Text
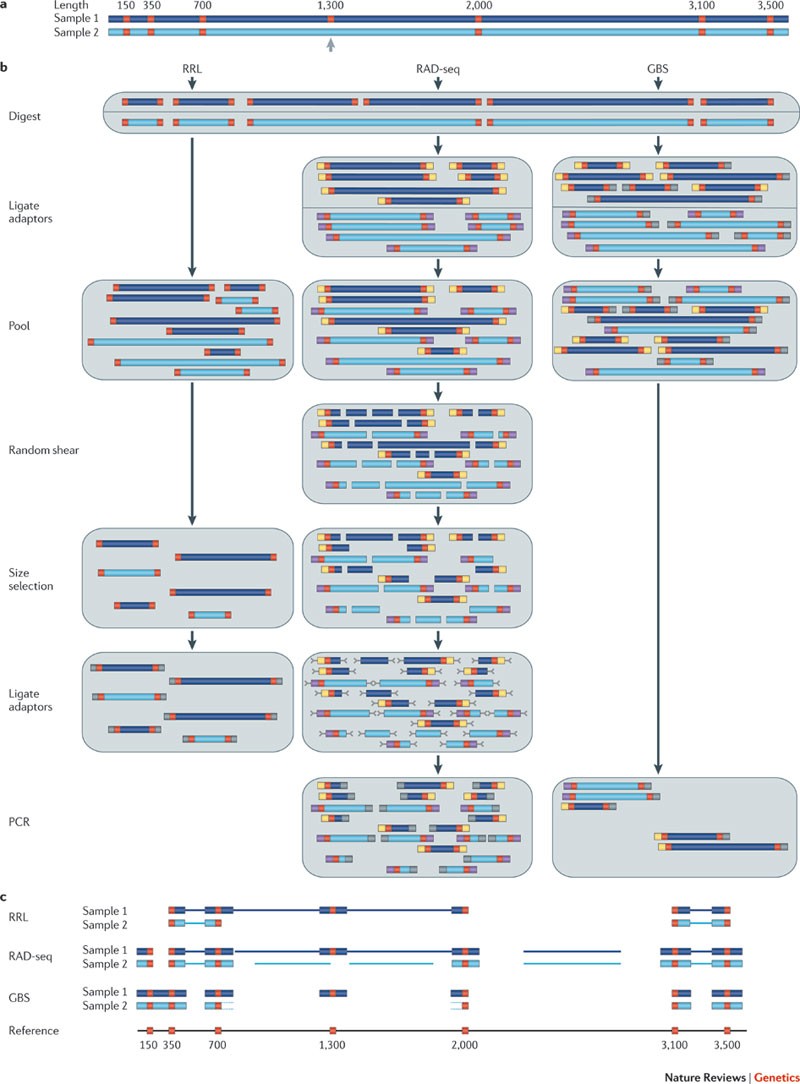
Genome-wide genetic marker discovery and genotyping using next-generation sequencing
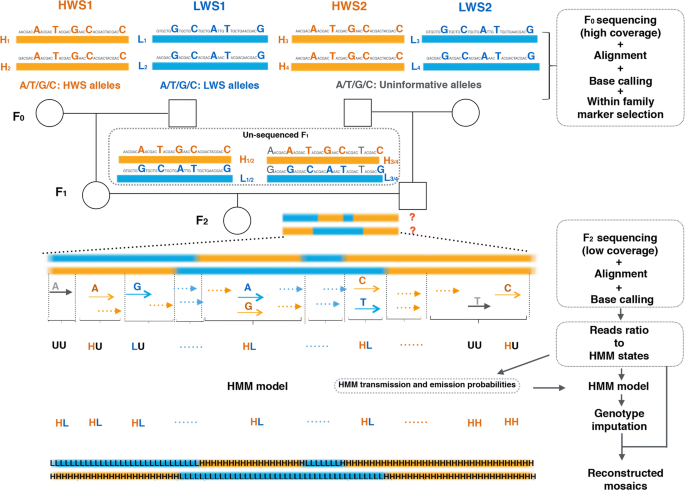
Genotyping by low-coverage whole-genome sequencing in intercross pedigrees from outbred founders: a cost-efficient approach, Genetics Selection Evolution
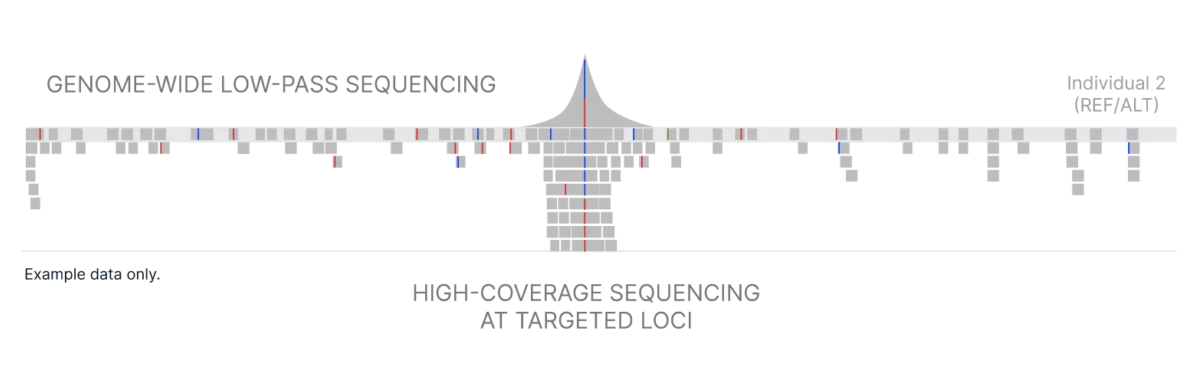
Introducing InfiniSEEK - Low-pass Genotyping Plus Target Capture in One Assay - Gencove

Phenotypic traits analyzed in the F 2 intercross.

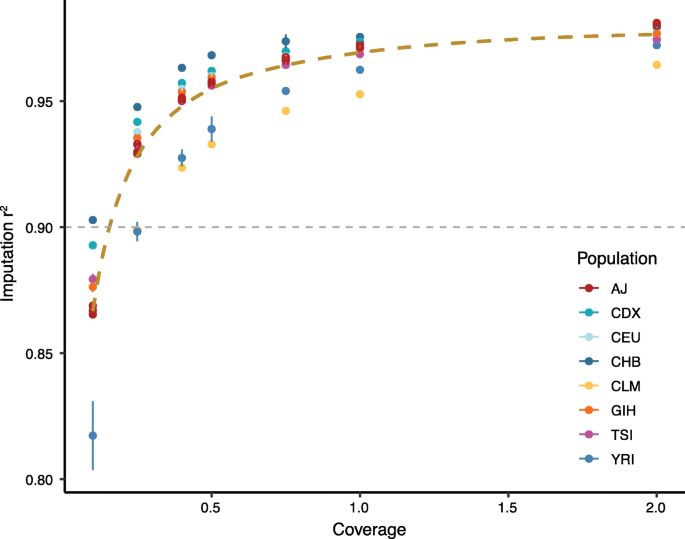
PDF] Accurate Genotype Imputation in Multiparental Populations from Low-Coverage Sequence
A Modern Approach to Genotyping for Plant and Animal Breeding - Gencove

Representative region of hemizygous deletion and a de novo CNV on

Combining genome‐wide association study based on low‐coverage whole genome sequencing and transcriptome analysis to reveal the key candidate genes affecting meat color in pigs - Zha - 2023 - Animal Genetics

Predictive ability for genomic prediction of traits using whole-genome

The minor allele frequency (x-axis distribution of collapsed SNPs using

Number of candidate de novo SNV site among four different depth of